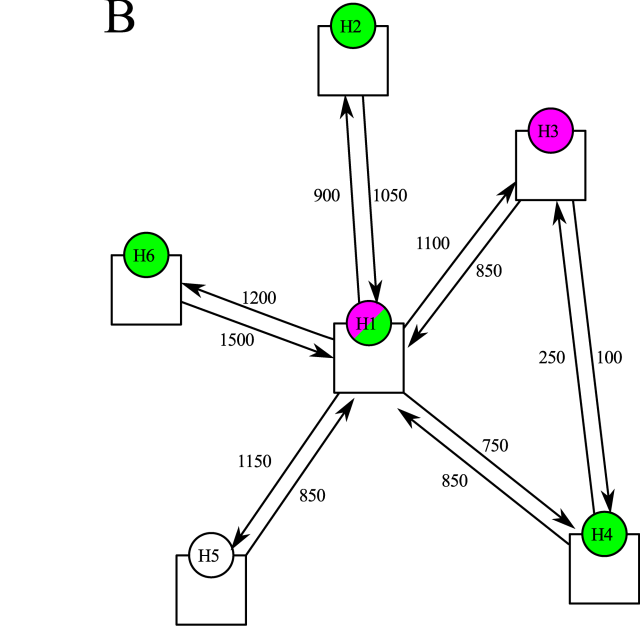
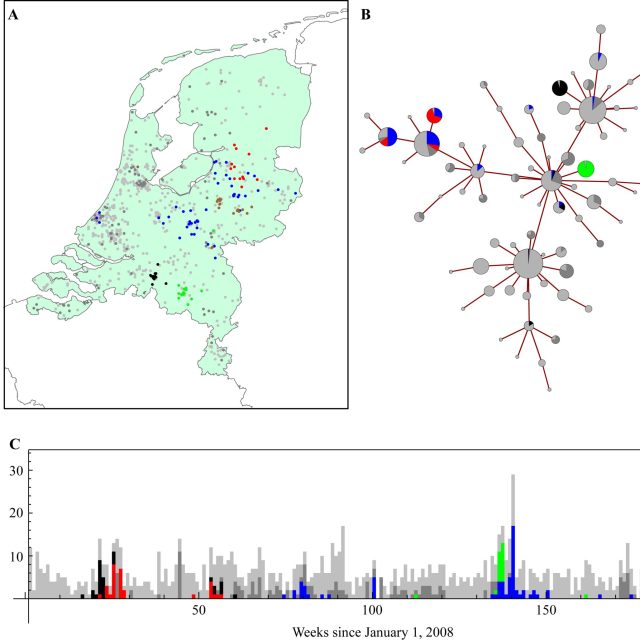
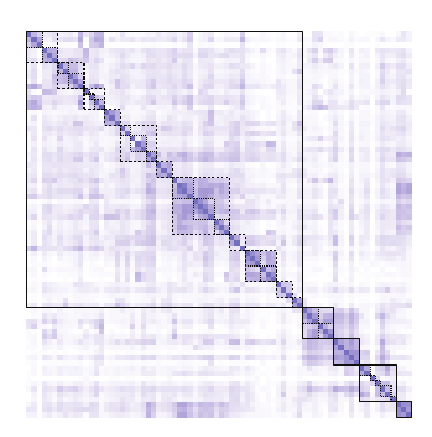
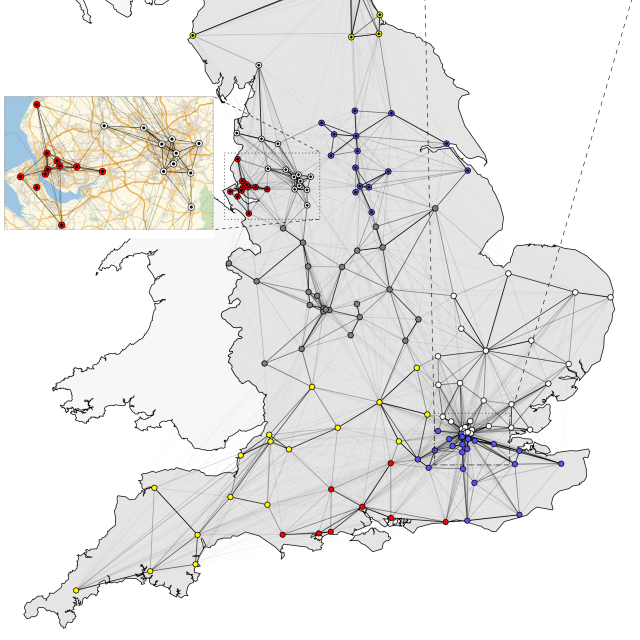
Siebler L, Rathje T, Calandri M, Stergiaropoulos K, Donker T, et al. A coupled experimental and statistical approach for an assessment of SARS-CoV-2 infection risk at indoor event locations. BMC Public Health 2023;23 (1), 1394
Donker T, The dangers of using large language models for peer review. The Lancet Infectious Diseases 2023;23 (7), 781
Scheithauer S, Dilthey A, Bludau A, Ciesek S, Corman V, Donker T, et al. Etablierung der Genomischen Erreger-Surveillance zur Stärkung des Pandemie-und Infektionsschutzes in Deutschland. Bundesgesundheitsblatt-Gesundheitsforschung-Gesundheitsschutz 2023;66 (4), 443-449
Budgell E, Davies T, Donker T, Hopkins S, Wyllie D, et al., Impact of hospital antibiotic use on patient-level risk of death among 36,124,372 acute and medical admissions in England. Journal of Infection 2022;84 (3)
Donker T, Bürkin FM, Wolkewitz M, Haverkamp C, Christoffel D, et al. Navigating hospitals safely through the COVID-19 epidemic tide: predicting case load for adjusting bed capacity. Infection Control & Hospital Epidemiology 2021;42 (6), 653-658
Donker T. Modelling how antimicrobial resistance spreads between wards. Elife 2020;9, e64228
Knight GM, Davies NG, Colijn C, Coll F, Donker T, et al. Mathematical modelling for antibiotic resistance control policy: do we know enough? BMC infectious diseases 2019;19, 1-9
Donker T, Smieszek T, Henderson KL, Walker TM, Hope R, et al. Using hospital network-based surveillance for antimicrobial resistance as a more robust alternative to self-reporting. Plos one 2019;14 (7), e0219994
Donker T, Henderson KL, Hopkins KL, Dodgson AR, Thomas S, et al.The relative importance of large problems far away versus small problems closer to home: insights into limiting the spread of antimicrobial resistance in England. BMC Medicine. 2017;15(1):86
Donker T, Smieszek T, Henderson KL, Johnson AP, Walker AS, Robotham JV. Measuring distance through dense weighted networks: The case of hospital-associated pathogens. PLoS Computational Biology. 2017;13(8):e1005622
Donker T, Reuter S, Scriberras J, Reynolds R, Brown NM, et al. Population genetic structuring of methicillin-resistant Staphylococcus aureus clone EMRSA-15 within UK reflects patient referral patterns. Microbial Genomics. 2017;3(7).
Chang H-H, Dordel J, Donker T, Worby CJ, Feil EJ, et al. Identifying the effect of patient sharing on between-hospital genetic differentiation of methicillin-resistant Staphylococcus aureus. Genome Medicine. 2016;8(1).
Donker T, Bosch T, Ypma RJF, Haenen APJ, van Ballegooijen WM, et al. Monitoring the spread of meticillin-resistant Staphylococcus aureus in The Netherlands from a reference laboratory perspective. Journal of Hospital Infection. 2016;93(4):366-374
Reuter S, Török ME, Holden MTG, Reynolds R, Raven KE, et al. Building a genomic framework for prospective MRSA surveillance in the United Kingdom and the Republic of Ireland. Genome Research. 2016;26(2):263-270
Yan X, Schouls LM, Pluister GN, Tao X, Yu X, et al. The population structure of Staphylococcus aureus in China and Europe assessed by multiple-locus variable number tandem repeat analysis; clues to geographical origins of emergence and dissemination. Clinical Microbiology and Infection. 2016;22(1)
Donker T, Ciccolini M, Wallinga J, Kluytmans JA, Grundmann H, Friedrich AW. Analyse van patiëntstromen: De basis voor regionale bestrijding van gevaarlijke infecties [Analysis of patient flows: basis for regional control of antibiotic resistance]. Nederlands tijdschrift voor geneeskunde. 2015; 159
Ciccolini M, Donker T, Grundmann H, Bonten MJM, Woolhouse MEJ. Efficient surveillance for healthcare-associated infections spreading between hospitals. Proceedings of the National Academy of Sciences. 2014 Jan 27;1–6.
Donker T, Wallinga J, Grundmann H. Dispersal of antibiotic-resistant high-risk clones by hospital networks: changing the patient direction can make all the difference. Journal of Hospital Infection. 2014 Jan;86(1):34–41.
Glasner C, Albiger B, Buist G, et al. Carbapenemase-producing Enterobacteriaceae in Europe: a survey among national experts from 39 countries, February 2013. Euro surveillance. 2013 July;18(28):1–7.
Ypma RJF, Donker T, van Ballegooijen WM, Wallinga J. Finding Evidence for Local Transmission of Contagious Disease in Molecular Epidemiological Datasets. PLoS ONE. 2013 Jul 26;8(7):e69875.
Ciccolini M, Donker T, Köck R, et al. Infection prevention in a connected world: The case for a regional approach. International Journal of Medical Microbiology; 2013 Mar;
Te Beest DE, Wallinga J, Donker T, Van Boven M (2013) Estimating the Generation Interval of Influenza A (H1N1) in a Range of Social Settings Epidemiology 24(2): 244-250. doi:10.1097/EDE.0b013e31827f50e8.
Greenland K, Whelan J, Fanoy E, Borgert M, Hulshof K, Yap K-B, Swaan C, Donker T, van Binnendijk R, de Melker H, Hahné S (2012) Mumps outbreak among vaccinated university students associated with a large party, the Netherlands, 2010 Vaccine 30(31): 4676–4680. doi:10.1016/j.vaccine.2012.04.083
Donker T, Wallinga J, Slack R, Grundmann H (2012) Hospital Networks and the Dispersal of Hospital-Acquired Pathogens by Patient Transfer PLoS ONE 7(4): e35002. doi:10.1371/journal.pone.0035002
Donker T, van Boven M, van Ballegooijen WM, van 't Klooster TM, Wielders CC, Wallinga J (2011) Nowcasting pandemic influenza A/H1N1 2009 hospitalizations in the Netherlands European Journal of Epidemiology 26(3): 195-201. doi:10.1007/s10654-011-9566-5
van Boven M, Donker T, van der Lubben M, van Gageldonk-Lafeber RB, te Beest DE, et al. (2010) Transmission of Novel Influenza A(H1N1) in Households with Post-Exposure Antiviral Prophylaxis. PLoS ONE 5(7): e11442. doi:10.1371/journal.pone.0011442
Donker T, Wallinga J, Grundmann H (2010) Patient Referral Patterns and the Spread of Hospital-Acquired Infections through National Health Care Networks. PLoS Comput Biol 6(3): e1000715. doi:10.1371/journal.pcbi.1000715
Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, et al. (2010) Geographic Distribution of Staphylococcus aureus Causing Invasive Infections in Europe: A Molecular-Epidemiological Analysis. PLoS Med 7(1): e1000215. doi:10.1371/journal.pmed.1000215
van 't Klooster TM, Wielders CC, Donker T, Isken L, Meijer A, van den Wijngaard CC, van der Sande MA, van der Hoek W (2010) Surveillance of Hospitalisations for 2009 Pandemic Influenza A(H1N1) in the Netherlands, 5 June - 31 December 2009. Euro Surveill. 15(2):pii=19461
Vriend H, Hahné S, Donker T, Meijer A, Timen A, van Steenbergen J, Osterhaus A, van der Sande M, Koopmans M, Wallinga J, Coutinho R, het Nederlandse nieuwe influenza A(H1N1)-onderzoeksteam (2009) De nieuwe influenza A(H1N1)-epidemie in Nederland. NTVG Vol. 153, A969
Hahné S, Donker T, Meijer A, Timen A, van Steenbergen J, Osterhaus A, van der Sande M, Koopmans M, Wallinga J, Coutinho R, the Dutch New Influenza A(H1N1)v Investigation Team. Epidemiology and control of influenza A(H1N1)v in the Netherlands: the first 115 cases. Euro Surveill. 2009;14(27):pii=19267